Tutorial 3#
February 27, 2025#
In the last two tutorials, you’ve taken your first steps in PyTorch and trained your first (convolutional) neural networks. You know now what convolution and cross-correlation do and what the essential steps in training a network are.
This week, you will train convolutional neural networks for 2D image segmentation. Keep in mind that everything you do and learn here can also be easily applied to 3D images. The data that we will be using today consists of chest X-ray images. The end goal in this dataset is to segment the ribs in these images, as in the image below. However, instead of segmenting and labeling all individual ribs, we will start a bit simpler and just segment every pixel that is a rib.

First, we set the data path. You can get the required images by downloading ribs.zip from the folder “Tutorial 3” here and uploading the folder to your coding environment.
Direct download
You can also directly download the file to your programming environment. If you are using the UT Jupyter server (as most of you are), open a terminal window on the server and run
wget -O ribs.zip https://surfdrive.surf.nl/files/index.php/s/Y4psc2pQnfkJuoT/download. This will download the file ribs.zip. Then, unzip this file by running unzip ribs.zip and you should find all image files on the server.
Set the data path to the “ribs” folder in the package that you have just downloaded in the code block beneath. If you’re running this on Colab, don’t forget to set a GPU runtime!
# ⌨️ add path:
data_path = "../data/Tutorial_3/ribs"
# check if data_path exists:
import os
if not os.path.exists(data_path):
print("Please update your data path to an existing folder.")
elif not set(["train", "val", "test"]).issubset(set(os.listdir(data_path))):
print("Please update your data path to the correct folder (should contain train, val and test folders).")
else:
print("Congrats! You selected the correct folder :)")
# Install additional packages required for this tutorial
!pip install scikit-image
!pip install wandb
!pip install monai

Weights and Biases#
In the previous tutorial, you have trained a neural network and monitored the loss curves in a Jupyter notebook. You can imagine that if you train multiple networks with different settings, it’s easy to lose track of all loss curves and figure out which model is best. Luckily, there exist excellent so-called MLOps systems to keep track of your experiments. Examples are Tensorboard, Neptune, and Weights & Biases (wandb). Here, we will give you some tips on how to use this last one to keep track of your experiments. Setting everything up is a simple process:
Register an account at https://wandb.ai/site. Your projects and runs will be shown on this website, which is accessible from anywhere.
Run the cell below (including
wandb.login()) to sign into your account. You will have to paste an API key, which you can find at https://wandb.ai/authorize, and hit Enter.
import wandb
wandb.login()
Now, we can initialize a run in wandb with wandb.init(). You can specify the project and run name, as well as configuration settings, by passing these as additional arguments.
run = wandb.init(project='Example project', name='Example run', config={'dataset': 'VinDr-RibCXR'})
By running the code above, a new run is created. Although we have not logged any information yet, it is possible to see the run on the wandb website. As soon as we will start logging information, it will become visible. It is possible to log information to your run with wandb.log(). When it’s time to complete the run, you can stop it with run.finish(). Run the cell below and see what happens on the run webpage.
import random
import time
# time.sleep(1) makes the script wait for 1 second, so the full script takes about 60 seconds to finish
for step in range(60):
wandb.log({'Loss': random.random(), 'Accuracy': random.random()})
time.sleep(1)
run.finish()
In the remainder of this notebook, we will use wandb to log our loss values to a run webpage. In this way, it is easier to keep track of multiple losses and to compare several runs.
Segmentation of chest X-ray images#
In this part of the tutorial, you will train a convolutional neural network for automatic segmentation of ribs in a chest X-ray image. You will examine the effects of using different loss functions, data augmentations and network architectures. To keep track of all these parameters, we will use wandb.
Data management#
Build a CacheDataset#
At the beginning of the notebook, you defined the path to the rib data. We will now use it to find and load the corresponding images. The data consists of an image and a label. However, the label is in this case not a binary label, but a segmentation mask. Both the segmentation masks and the X-rays are .png files and can be opened using the PIL library.
For this tutorial we are not going to code the Dataset class ourselves but instead use a Monai class: CacheDataset. It has two main advantages:
as the name suggests, it uses a cache mechanism, which means that it is memory efficient (this is partly explained later).
everything is pre-built, so no need to code the
__init__,__getitem__and__len__functions anymore.
CacheDataset only needs the list of your data samples to work. As we are learning a segmentation task, one data sample is composed of two images: the chest X-ray (named img) and the mask of the ribs (named mask). With Python, we represent this data sample with a dictionary:

Then we could give this kind of list to CacheDataset to create our data set:

However, as this list contains all the images, it means that all the images are always in Python memory even though they are not currently used so this is very inefficient. This is why we don’t directly give the images to CacheDataset, but the paths to find them on our disk. Then we also need to give a Transform object, LoadRibData which will transform these paths into the corresponding images only when they are needed.


As we don’t want to write this list of paths manually, we write the function build_dict_ribs which automatically computes it based on the root folder of the data directory data_path. This is possible because fortunately your teachers pre-organized the dataset for this tutorial, but be aware that most medical datasets require a lot of cleaning up prior to this.
import os
import numpy as np
import matplotlib.pyplot as plt
import glob
import monai
from PIL import Image
import torch
def build_dict_ribs(data_path, mode='train'):
"""
This function returns a list of dictionaries, each dictionary containing the keys 'img' and 'mask'
that returns the path to the corresponding image.
Args:
data_path (str): path to the root folder of the data set.
mode (str): subset used. Must correspond to 'train', 'val' or 'test'.
Returns:
(List[Dict[str, str]]) list of the dictionaries containing the paths of X-ray images and masks.
"""
# test if mode is correct
if mode not in ["train", "val", "test"]:
raise ValueError(f"Please choose a mode in ['train', 'val', 'test']. Current mode is {mode}.")
# define empty dictionary
dicts = []
# list all .png files in directory, including the path
paths_xray = glob.glob(os.path.join(data_path, mode, 'img', '*.png'))
# make a corresponding list for all the mask files
for xray_path in paths_xray:
if mode == 'test':
suffix = 'val'
else:
suffix = mode
# find the binary mask that belongs to the original image, based on indexing in the filename
image_index = os.path.split(xray_path)[1].split('_')[-1].split('.')[0]
# define path to mask file based on this index and add to list of mask paths
mask_path = os.path.join(data_path, mode, 'mask', f'VinDr_RibCXR_{suffix}_{image_index}.png')
if os.path.exists(mask_path):
dicts.append({'img': xray_path, 'mask': mask_path})
return dicts
class LoadRibData(monai.transforms.Transform):
"""
This custom Monai transform loads the data from the rib segmentation dataset.
Defining a custom transform is simple; just overwrite the __init__ function and __call__ function.
"""
def __init__(self, keys=None):
pass
def __call__(self, sample):
image = Image.open(sample['img']).convert('L') # import as grayscale image
image = np.array(image, dtype=np.uint8)
mask = Image.open(sample['mask']).convert('L') # import as grayscale image
mask = np.array(mask, dtype=np.uint8)
# mask has value 255 on rib pixels. Convert to binary array
mask[np.where(mask==255)] = 1
return {'img': image, 'mask': mask, 'img_meta_dict': {'affine': np.eye(2)},
'mask_meta_dict': {'affine': np.eye(2)}}
Note
Note that LoadRibData is now outputing a more complex dictionary with two additional keys: img_meta_dict and mask_meta_dict. These contain respectively the meta data of img and mask: here we decided to add the resolution of the image, corresponding to the key affine. This will be useful for Monai to perform some transformations later.
A CacheDataset needs two arguments to perform our procedure
The list of samples (represented by dictionaries) which is computed by
build_dict_ribs.A transform object,
LoadRibData, which will transform the paths in the previous list into images.
# construct list of dictionaries
train_dict_list = build_dict_ribs(data_path, mode='train')
# construct CacheDataset from list of paths + transform
train_dataset = monai.data.CacheDataset(train_dict_list, transform=LoadRibData())
Exercise
How many samples are in the training, validation, and test set?
Answer key
val_dict_list = build_dict_ribs(data_path, mode='val')
test_dict_list = build_dict_ribs(data_path, mode='test')
val_dataset = monai.data.CacheDataset(val_dict_list, transform=LoadRibData())
test_dataset = monai.data.CacheDataset(test_dict_list, transform=LoadRibData())
print(f'{train_dataset.__len__()=}')
print(f'{val_dataset.__len__()=}')
print(f'{test_dataset.__len__()=}')
Exercise
What are the dimensions of the images? Are they all equal? Will this a problem when training a CNN for segmentation?
Answer key
for i in range(5):
sample = train_dataset[i]
img = sample['img']
mask = sample['mask']
print(f'{img.shape=}, {mask.shape=}')
They are different, which means that we can’t stack them in mini-batches.
Exercise
What is the pixel intensity range of the images? Are they similar? Will this be a problem when training a CNN for segmentation?
Visualize the pixel intensity range of a bunch of images. Hint Use Matplotlib hist (documentation) and .flatten() your image (or you will get >2000 histograms per image)
Answer key
bins = np.linspace(0, 255, 50)
for i in range(5):
sample = train_dataset[i]
img = sample['img']
img = img.flatten()
plt.hist(img, bins, alpha=0.5, label=i)
plt.show()
Image visualization#
We now build visualize_rib_sample to plot X-ray images on which we overlay their binary mask (in green).
def visualize_rib_sample(sample, title=None):
# Visualize the x-ray and overlay the mask, using the dictionary as input
image = np.squeeze(sample['img'])
mask = np.squeeze(sample['mask'])
plt.figure(figsize=[10,7])
plt.imshow(image, 'gray')
overlay_mask = np.ma.masked_where(mask == 0, mask == 1)
plt.imshow(overlay_mask, 'Greens', alpha = 0.7, clim=[0,1], interpolation='nearest')
if title is not None:
plt.title(title)
plt.show()
Exercise
Use visualize_rib_sample to visualize samples from the training set.
Answer key
sample_dict = train_dataset[0]
visualize_rib_sample(sample_dict)
Data transforms#
The training dataset that you have created in the previous step is quite small. In order to improve the generalization of the network, we are going to do some data augmentation. This can be easily implemented in the pipeline using the transforms in Monai.
Data augmentation
Data augmentation is a very common way to enlarge the number of training samples that you use. By applying transformations such as rotation, flipping, scaling to images we artificially enlarge the number of samples that the model sees.
Monai distinguishes deterministic and random transforms:
Deterministic transforms do the exact same thing every time they are called, for example intensity normalization will always have the same outcome.
Random transforms can be rotations or flips that do not have the same outcome each time they are called.
The random transforms (for example flipping and rotation) need to be performed in the same way on the input image 'img' as well as on the label 'mask'. Monai has adapted transforms that take dictionaries as an input, such that random transforms can be performed in the same way for both images: you don’t want a different angle of rotation for the image and the mask, as they will become misaligned! You can recognize these transforms as they end with a ‘d’, e.g. Zoomd instead of Zoom.

The examples below show how to use one of these dictionary transforms to perform a random horizontal flip and a random rotation. The procedure of applying a transform from Monai consists of two steps:
Initialize the transform: here you build the object and pass the init variables, such as the probability or range of rotation, and in case of a dictionary transform the keys of the dictionary.
Apply the transform on a data sample
Lastly, most transforms expect a channel dimension for both 2D and 3D images. As we have only one channel, we should add this extra dimension using the monai.transforms.AddChanneld transform first.
# Load sample
index = np.random.choice(np.arange(len(train_dataset))) # This picks a random sample, but you can change this value
sample_dict = train_dataset[index]
visualize_rib_sample(sample_dict, title="Original sample")
# Add channels
add_channels_transform = monai.transforms.AddChanneld(keys=['img', 'mask']) # Initialize the transform
channels_sample_dict = add_channels_transform(sample_dict) # Apply the transform
print("Size of the image before AddChanneld transform", sample_dict["img"].shape)
print("Size of the image after AddChanneld transform", channels_sample_dict["img"].shape)
# Random flip
# here we define the keys, the probability that the flip is performed and the axis to flip over
random_flip_transform = monai.transforms.RandFlipd(keys=['img', 'mask'], prob=1, spatial_axis=1)
# We put a probability of 1 to always flip the image for visualization purposes.
# Please DO NOT DO THAT in the rest of the notebook.
flipped_sample_dict = random_flip_transform(channels_sample_dict)
visualize_rib_sample(flipped_sample_dict, title="(Not quite randomly) flipped sample")
# Random rotation
# Here we define the keys in the dictionary that contain the data, the rotation range, but also the interpolation mode.
# The interpolation mode defines how new pixel values for the rotated image are computed.
# Note that these differ between mask and image, as we want to keep binary labels for the masks, and (bi)linear interpolation
# would result in scalar values between 0 and 1.
random_rotation_transform = monai.transforms.RandRotated(keys=['img', 'mask'], range_x=np.pi/4, prob=1, mode=['bilinear', 'nearest'])
rotated_sample_dict = random_rotation_transform(channels_sample_dict)
visualize_rib_sample(rotated_sample_dict, title="Randomly rotated sample")
Note how we concatenated the AddChannelsd transform with the flip and rotate transform by applying them after each other. If we have only two transforms, like here, this is an acceptable workflow. However, if we have a whole bunch of transforms that we have to call during training of the network, you can imagine that the code will become very messy. Therefore, we can combine multiple transforms into a single one using monai.transforms.Compose.
Upon initialization, Compose takes a list of transforms as input and outputs a transform object that concatenates all the transforms. It can be applied in the same way as the transforms shown above.
Exercise
Use the Compose class of Monai to compose AddChanneld, RandFlipd and RandRotated in a single transform. Then apply this composition to a data set sample.
Answer key
sample_dict = train_dataset[0]
# Create the composed transform
add_channels_transform = monai.transforms.AddChanneld(keys=['img', 'mask']) # Initialize the transform
random_flip_transform = monai.transforms.RandFlipd(keys=['img', 'mask'], prob=1, spatial_axis=1)
random_rotation_transform = monai.transforms.RandRotated(keys=['img', 'mask'], range_x=np.pi/4, prob=1, mode=['bilinear', 'nearest'])
transforms = monai.transforms.Compose([
add_channels_transform,
random_flip_transform,
random_rotation_transform
])
# Apply this new single transform to sample_dict
transformed_sample = transforms(sample_dict)
visualize_rib_sample(transformed_sample, title="Transformed sample")
Efficiency
As mentioned before, there is a distinction between random and deterministic transforms. This distinction is important, as their order can have a huge influence on computational efficiency. The CacheDataset class stores the outcomes of all deterministic transforms in memory, and performs the random transforms on-the-fly. Therefore, placing the deterministic transforms before the random ones in combination with caching results in a training and inference procedure will result in higher computational efficiency.
We are going to use the following three transforms in the X-ray segmentation task
ZoomRandomFlipRandSpatialCrop
Exercise
Compose the AddChannel, ScaleIntensity, Zoom, RandomFlip and RandSpatialCrop transforms in a computationally efficient order and construct a new training dataset (with CacheDataset) using these transforms.
Make sure to visualize some samples to see if the transforms are working as you expect.
Don’t forget to put
LoadRibDataas your first transform or your dataset will try to rotate strings representing the paths of your images…If you identified problems in the previous sections it is time to solve them!
Answer key
train_transform = monai.transforms.Compose([
LoadRibData(),
monai.transforms.AddChanneld(keys=['img', 'mask']),
monai.transforms.ScaleIntensityd(keys=['img'],minv=0, maxv=1),
monai.transforms.Zoomd(keys=['img', 'mask'], zoom=0.25, keep_size=False, mode=['bilinear', 'nearest']),
monai.transforms.RandFlipd(keys=['img', 'mask'], prob=0.5, spatial_axis=1),
monai.transforms.RandSpatialCropd(keys=['img', 'mask'], roi_size=[256,256], random_size=False)
])
train_dataset = monai.data.CacheDataset(train_dict_list, transform=train_transform)
for i in range(5):
visualize_rib_sample(train_dataset[i], title=f"Transformed sample {i}")
Dataloader#
We use mini-batch gradient descent during training, so we want to sample batches to train the network on, rather than single instances of the data. For this we use monai.data.DataLoader, which efficiently samples batches from the data that can be fed into the network right away. Remember that you have also used a DataLoader in Tutorial 2.
This PyTorch tutorial provides more information about datasets and dataloaders.
Exercise
Construct a dataloader with randomly sampled mini-batches of 16 images for the training set.
Answer key
train_loader = monai.data.DataLoader(train_dataset, batch_size=16, shuffle=True)
print(next(iter(train_loader))['img'].shape)
Validation set#
During training, we want to keep an eye on how the network generalizes to unseen data. It could perform very well on its training set, whereas it performs poorly on unseen data (overfitting). For this, we need to construct a validation set consisting of comparable but different samples of data than the training set. For our data, it can be constructed in a similar way as the train set:
Construct the dictionary of file paths
Define the transforms that should be applied on the validation data
Construct the CacheDataset using the dictionary and transform
Build a validation dataloader using from the CacheDataset
In contrast to the training set, we don’t apply data augmentation to the validation set. This is important to keep in mind when defining transforms.
Exercise
Build a validation dataset and dataloader using these steps. Create a separate validation transform.
Answer key
validation_dict = build_dict_ribs(data_path, mode='val')
validation_transforms = monai.transforms.Compose([
LoadRibData(),
monai.transforms.AddChanneld(keys=['img', 'mask']),
monai.transforms.ScaleIntensityd(keys=['img'],minv=0, maxv=1),
monai.transforms.Resized(keys=['img', 'mask'], spatial_size=[256,256])
])
validation_data = monai.data.CacheDataset(validation_dict, transform=validation_transforms)
validation_loader = monai.data.DataLoader(validation_data, batch_size=16)
print(next(iter(validation_loader))['img'].shape)
Setting up the neural network, loss function, and optimizer#
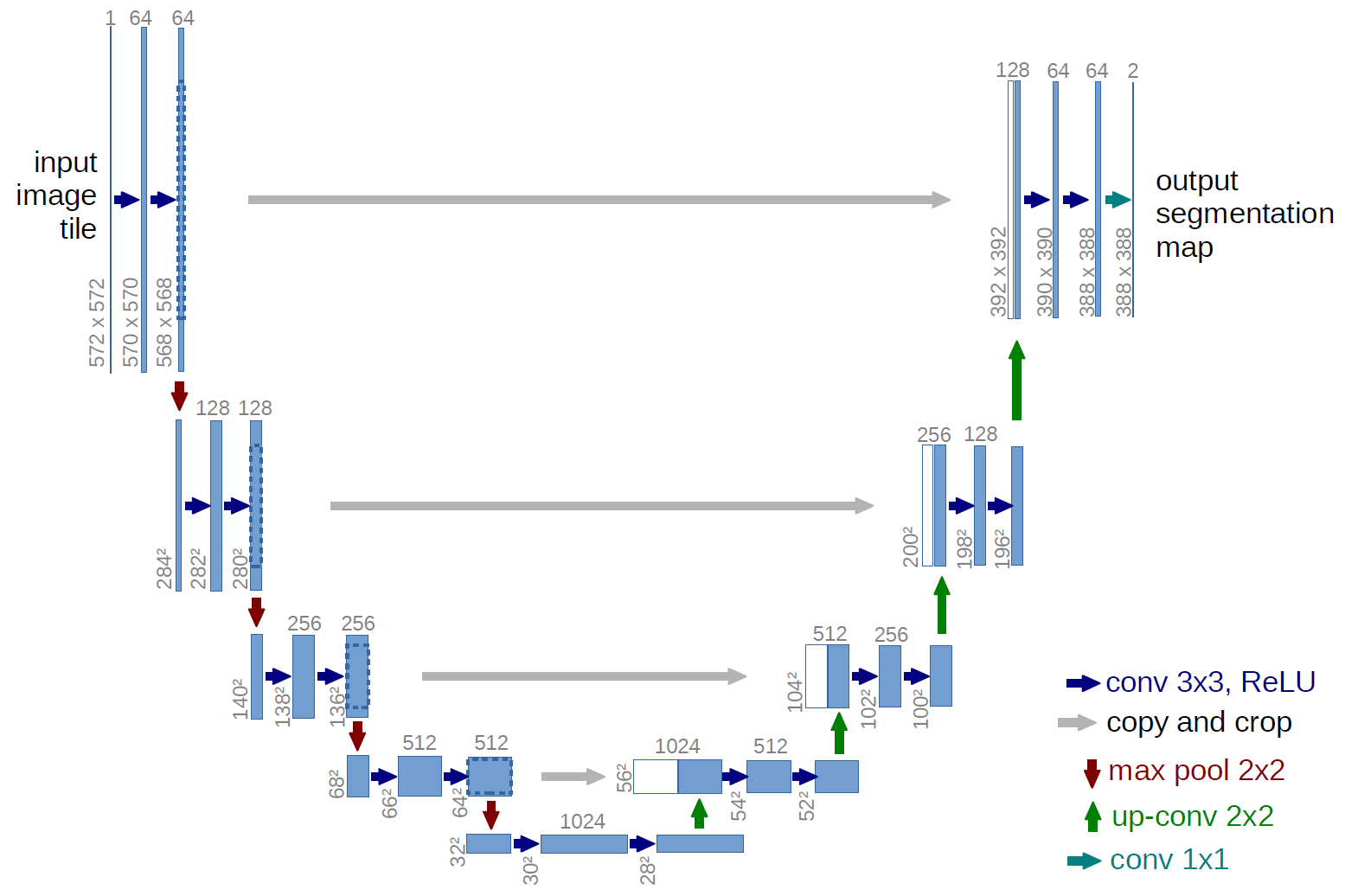
Now that we have the data loading process out of the way, we set up a U-Net, a loss function, and an optimizer. If possible, use a GPU to substantially speed up computing.
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f'The used device is {device}')
Setting up a U-Net for segmentation in MONAI is as easy as calling the UNet function and providing it with the number of input channels, output channels, and feature maps/channels in the intermediate layers. The following provides us with a model that is optimized during training to perform segmentation.
model = monai.networks.nets.UNet(
spatial_dims=2,
in_channels=1,
out_channels=1,
channels=(8, 16, 32, 64, 128),
strides=(2, 2, 2, 2),
num_res_units=2,
).to(device)
Exercise
How many levels does the U-Net have? And how many parameters does it have (use the code from last tutorial)?
Answer key
num_params = sum(p.numel() for p in model.parameters() if p.requires_grad)
print(f'U-Net has 5 layers (see "channels" parameter) and {num_params} parameters')
Loss function#
The loss function should reflect what we want the training model to be able to do. For segmentation, a popular function to use for training the network is the Dice loss, which measures the overlap between the ground-truth and the model prediction. Monai offers a wide range of different loss functions that are also suitable for segmentation. In this tutorial, we will also assess the effects of using different loss functions on our network performance.
We show how to implement the Dice function in this example. Our network only has one output channel. In order to have all the output values between 0 and 1, so we can compute the Dice, the function first applies a logistic sigmoid. The Dice loss is computed over the full mini-batch (batch=True) to avoid poorly defined loss in individual batch samples. This means that, in a sense, we compute the Dice loss in a 3D stack of 2D images.
loss_function = monai.losses.DiceLoss(sigmoid=True, batch=True)
Choose an optimizer#
An optimizer algorithm is chosen that performs gradient descent on the network parameters to minimize the loss function. Last week you have used SGD (stochastic gradient descent). In many cases, Adam is a good default option. The optimizer operates on the parameters of the previously defined U-Net, i.e. model. A learning rate lr is provided to the optimizer, which defines how large the changes should be that are made to the network parameters in each iteration.
optimizer = torch.optim.Adam(model.parameters(), lr=1e-3)
Setting up the training loop#
In the previous tutorial, we explained how to set up a simple loop for training your network. For training this network, the loop can be set up in a similar way. Instead of keeping track of the train and validation loss on your computer, you can log everything in weights and biases.
Exercise
Set up a simple loop for training the network and log the following in weights and biases:
Loss function
Learning rate
Training loss
Validation loss
Some validation images, including segmentation masks that display model output and the ground truth (see W&B documentation).
Below, we provide a log_to_wandb() function that you could use for this. Just call it once at the end of every epoch, with the right arguments.
Answer key
from tqdm import tqdm
import wandb
run = wandb.init(
project='tutorial3_segmentation',
name='test',
config={
'loss function': str(loss_function),
'lr': optimizer.param_groups[0]["lr"],
'transform': from_compose_to_list(train_transform),
'batch_size': train_loader.batch_size,
}
)
# Do not hesitate to enrich this list of settings to be able to correctly keep track of your experiments!
# For example you should add information on your model...
run_id = run.id # We remember here the run ID to be able to write the evaluation metrics
def wandb_masks(mask_output, mask_gt):
""" Function that generates a mask dictionary in format that W&B requires """
# Apply sigmoid to model ouput and round to nearest integer (0 or 1)
sigmoid = torch.nn.Sigmoid()
mask_output = sigmoid(mask_output)
mask_output = torch.round(mask_output)
# Transform masks to numpy arrays on CPU
# Note: .squeeze() removes all dimensions with a size of 1 (here, it makes the tensors 2-dimensional)
# Note: .detach() removes a tensor from the computational graph to prevent gradient computation for it
mask_output = mask_output.squeeze().detach().cpu().numpy()
mask_gt = mask_gt.squeeze().detach().cpu().numpy()
# Create mask dictionary with class label and insert masks
class_labels = {1: 'ribs'}
masks = {
'predictions': {'mask_data': mask_output, 'class_labels': class_labels},
'ground truth': {'mask_data': mask_gt, 'class_labels': class_labels}
}
return masks
def log_to_wandb(epoch, train_loss, val_loss, batch_data, outputs):
""" Function that logs ongoing training variables to W&B """
# Create list of images that have segmentation masks for model output and ground truth
log_imgs = [wandb.Image(img, masks=wandb_masks(mask_output, mask_gt)) for img, mask_output,
mask_gt in zip(batch_data['img'], outputs, batch_data['mask'])]
# Send epoch, losses and images to W&B
wandb.log({'epoch': epoch, 'train_loss': train_loss, 'val_loss': val_loss, 'results': log_imgs})
for epoch in tqdm(range(200)):
# training
model.train()
epoch_loss = 0
step = 0
for batch_data in train_loader:
step += 1
optimizer.zero_grad()
outputs = model(batch_data["img"].float().to(device))
loss = loss_function(outputs, batch_data["mask"].to(device))
loss.backward()
optimizer.step()
epoch_loss += loss.item()
train_loss = epoch_loss/step
# validation
step = 0
val_loss = 0
for batch_data in validation_loader:
step += 1
model.eval()
outputs = model(batch_data['img'].float().to(device))
loss = loss_function(outputs, batch_data['mask'].to(device))
val_loss+= loss.item()
val_loss = val_loss / step
log_to_wandb(epoch, train_loss, val_loss, batch_data, outputs)
# Store the network parameters
torch.save(model.state_dict(), r'trainedUNet.pt')
run.finish()
Evaluate the trained network#
Visual inspection#
We have trained the network on small patches of the image. In order to see how well it performs on the entire image, we can use the monai.inferers.SlidingWindowInferer. This ‘slides’ the network over patches of the input image. The overlap between patches can also be controlled.
Moreover, we want our final mask to have discrete values (0 or 1). Then, we need to discretize the continuous output of the network. This is why we use Sigmoid (clipping the output values between 0 and 1) and AsDiscrete (mapping the continuous distribution on the {0, 1} set).
def visual_evaluation(sample, model):
"""
Allow the visual inspection of one sample by plotting the X-ray image, the ground truth (green)
and the segmentation map produced by the network (red).
Args:
sample (Dict[str, torch.Tensor]): sample composed of an X-ray ('img') and a mask ('mask').
model (torch.nn.Module): trained model to evaluate.
"""
model.eval()
inferer = monai.inferers.SlidingWindowInferer(roi_size=[256, 256])
discrete_transform = monai.transforms.AsDiscrete(logit_thresh=0.5, threshold_values=True)
Sigmoid = torch.nn.Sigmoid()
with torch.no_grad():
output = discrete_transform(Sigmoid(inferer(sample['img'].to(device), network=model).cpu())).squeeze()
fig, ax = plt.subplots(1,3, figsize = [12, 10])
# Plot X-ray image
ax[0].imshow(sample["img"].squeeze(), 'gray')
ax[1].imshow(sample["img"].squeeze(), 'gray')
# Plot ground truth
mask = np.squeeze(sample['mask'])
overlay_mask = np.ma.masked_where(mask == 0, mask == 1)
ax[1].imshow(overlay_mask, 'Greens', alpha = 0.7, clim=[0,1], interpolation='nearest')
ax[1].set_title('Ground truth')
# Plot output
overlay_output = np.ma.masked_where(output < 0.1, output >0.99)
ax[2].imshow(sample['img'].squeeze(), 'gray')
ax[2].imshow(overlay_output, 'Reds', alpha = 0.7, clim=[0,1])
ax[2].set_title('Prediction')
plt.show()
test_dict = build_dict_ribs(data_path, mode='test')
test_transform = monai.transforms.Compose([
LoadRibData(),
monai.transforms.AddChanneld(keys=['img', 'mask']),
monai.transforms.ScaleIntensityd(keys=['img'],minv=0, maxv=1),
monai.transforms.Zoomd(keys=['img', 'mask'], zoom=0.25, keep_size=False, mode=['bilinear', 'nearest']),
]
)
test_set = monai.data.CacheDataset(test_dict, transform=test_transform)
test_loader = monai.data.DataLoader(test_set, batch_size=1)
for sample in test_loader:
visual_evaluation(sample, model)
Compute evaluation metrics#
Previously, we evaluated the quality of our segmentation on the test set by visually inspecting the quality of the maps produced by our network. Though this is an essential step when developping and debugging a network, this step is also quite subjective, and tedious when there are a lot of images. This is why we need metrics to evaluate the performance of our network! These metrics give us a quantity that represents the performance of our network and enable comparison to other methods.
We provide the function compute_metric to evaluate the performance of the network on the segmentation task. Similar to the transforms and loss functions, Monai contains many evaluation metrics to assess the model’s performance. We show how to compute the Dice metric for the network.
def compute_metric(dataloader, model, metric_fn):
"""
This function computes the average value of a metric for a data set.
Args:
dataloader (monai.data.DataLoader): dataloader wrapping the dataset to evaluate.
model (torch.nn.Module): trained model to evaluate.
metric_fn (function): function computing the metric value from two tensors:
- a batch of outputs,
- the corresponding batch of ground truth masks.
Returns:
(float) the mean value of the metric
"""
model.eval()
inferer = monai.inferers.SlidingWindowInferer(roi_size=[256, 256])
discrete_transform = monai.transforms.AsDiscrete(threshold=0.5)
Sigmoid = torch.nn.Sigmoid()
mean_value = 0
for sample in dataloader:
with torch.no_grad():
output = discrete_transform(Sigmoid(inferer(sample['img'].to(device), network=model).cpu()))
mean_value += metric_fn(output, sample["mask"])
return (mean_value / len(dataloader)).item()
As we want to log everything in the corresponding run with W&B, we access to the corresponding run with wandb.Api().
api = wandb.Api()
run = api.run(f"tutorial3_segmentation/{run_id}")
Dice#
This is the same metric that the one that was used for the loss function.
Exercise
Find the appropriate function in monai.metrics to compute the mean Dice with compute_metric.
Answer key
metric_fn = monai.metrics.compute_meandice
dice = compute_metric(test_loader, model, metric_fn)
run.summary["dice"] = dice
print(f"Dice on test set: {dice:.3f}")
Hausdorff distance#
The Hausdorff distance uses a function f which computes the minimal distance between the farthest point that can be found in a set Y compared to another set X. The Hausdorff takes the worst case between f(X,Y) and f(Y,X).

Exercise
Find the appropriate function in monai.metrics to compute the Hausdorff distance with compute_metric.
Answer key
metric_fn = monai.metrics.compute_hausdorff_distance
Hausdorff_dist = compute_metric(test_loader, model, metric_fn)
run.summary["Hausdorff_dist"] = Hausdorff_dist
print(f"Hausdorff distance on test set: {Hausdorff_dist:.3f}")
Exercise
Are you happy with the performance of your model? Can you think of ways to improve the performance?
Comparison to baseline masks#
Although you did some visual inspection of the outputs of the model, interpretation of the Dice scores and Hausdorff distances is not trivial. Therefore, comparing to a baseline model for which we know it performs poorly is a good way to get some idea of how good your model actually is.
For our dummy baseline model, instead of rib-shaped segmentations, we let the model output square segmentation maps in the rib region.
from skimage import measure
def make_dummy_sample(sample):
M = sample['mask'].squeeze()
labels = measure.label(M)
dummy_labels = np.zeros((labels.shape[0], labels.shape[1]))
for i in np.unique(labels):
if i > 0:
mask_locs = np.where(labels == i)
limits = [np.min(mask_locs[0]), np.max(mask_locs[0]), np.min(mask_locs[1]), np.max(mask_locs[1])]
dummy_labels[limits[0]:limits[1], limits[2]:limits[3]] = 1
return torch.tensor(dummy_labels)
sample = test_set[0]
visualize_rib_sample(sample, title = 'original mask')
visualize_rib_sample({'img': sample['img'], 'mask': make_dummy_sample(sample)}, title = 'dummy segmentation masks')
def compute_metric_dummy(dataloader, metric_fn):
"""
This function computes the average value of a metric for a data set using the dummy segmentation masks
Args:
dataloader (monai.data.DataLoader): dataloader wrapping the dataset to evaluate.
metric_fn (function): function computing the metric value from two tensors:
- a batch of outputs,
- the corresponding batch of ground truth masks.
Returns:
(float) the mean value of the metric
"""
mean_value = 0
A = monai.transforms.AddChannel()
for sample in dataloader:
output = make_dummy_sample(sample)
mean_value += metric_fn(A(A(output)), sample["mask"])
return (mean_value / len(dataloader)).item()
print(f'Mean Dice: {compute_metric_dummy(test_loader, monai.metrics.compute_meandice):.3f}')
Exercise
Are you still happy with the performance of your model?
Part 3: Multiclass classification#
Exercise
In the data directory, we have also placed a folder called mask_mc for the train, val and test set. In these images, we have labeled the left and right 7th rib. This is one of the ribs that’s most prone to fracture, and identifying exactly that rib and the side that it’s on could be valuable. Thus, this is no longer a binary label segmentation problem, but a multiclass segmentation problem. Try to also segment these images, consider what you will have to change in your code for this to work.
# ⌨️ code your answer here


